Frequency of HPV-16 in Saliva and Blood in Head and Neck Squamous Cell Carcinoma in Pakistani Population
OBJECTIVE: Pakistan is geographically located in the region where prevalence of head and neck squamous cell carcinoma (HNSCC) is high. Human papilloma virus (HPV) subtype-16 plays significant role in heterogeneity of HNSCC. The objective of this study was to evaluate the frequency of HPV-16 in both blood and saliva in HNSCC in our population and its association with clinicopathological features.
METHODOLOGY: A total of 200 samples comprising 100 blood and 100 saliva samples were collected. Hundred samples (saliva; n=50, blood; n=50) were collected from histopathologically confirmed head and neck squamous cell carcinoma patients and 100 samples (saliva; n=50 saliva, blood; n=50) were collected from healthy individuals without a history of cancer as controls. HPV-16 was detected using conventional polymerase chain reaction and gel-electrophoresis. The frequency and distribution of clinicopathological variables were calculated in percentages. Chi-square test was used to analyze the association of HPV-16 positivity with clinicopathological variables.
RESULTS: Of 100 samples from HNSCC patients, 11 (11%) samples were positive for HPV-16. Of 11 HPV-16 positive samples, 8 (16%) were detected in saliva (n=50) and 3 (6%) were detected in plasma (n=50). Of 100 samples from control group, both blood and saliva samples were negative for HPV-16. No statistically significant association was found between HPV-16 in saliva and blood and clinicopathological characteristics (P > 0.05).
CONCLUSION: Within the limit of current study, HPV-16 has a low-frequency in saliva and blood in HNSCC in our population. Further studies on HNSCC with large sample and HPV-subtypes are suggested. KEYWORD: Blood; human papilloma virus; saliva; squamous cell carcinoma.
HOW TO CITE: Saher S, Syed SA, Iqbal A, Syed FA, Amin F, Sultan A, Anees M. Frequency of HPV-16 in saliva and blood in head and neck squamous cell carcinoma in Pakistani population. J Pak Dent Assoc 2020;29(4):228-234.
DOI: https://doi.org/10.25301/JPDA.294.228
Received: 07 August 2020, Accepted: 03 September 2020
INTRODUCTION
Head and neck squamous cell carcinoma (HNSCC) is the seventh most common malignancy across the globe.1 It accounts for 3 % of all cancers and arises from epithelial lining of the oral and nasal cavity, pharynx, larynx, paranasal sinuses, salivary glands, and lymph nodes. Histopathologically, about 95 % of head and neck cancers are squamous cell carcinoma (SCC).2 Pakistan, a developing country with many health-related challenges to its population, is geographically located in the region where prevalence of HNSCC is high.3,4 Tobacco and alcohol are the major risk-factors that contribute to the development of HNSCC in developing countries. Emerging evidence suggests role of human papilloma virus (HPV) infection in the heterogeneity of HNSCC particularly in developed countries.5,6 Moreover, the prevalence of HPV-driven HNSCC varies geographically across the globe.7 The HPV-driven HNSCC differs significantly from the HPV negative HNSCC, in terms of the molecular, therapeutic and prognostic aspects. Therefore, it is essential to discriminate between HPV-positive and HPV-negative HNSCC.5 Recently, a systematic review reported a high prevalence of HPV subtype 16 in HNSCC in Bangladesh, India and Pakistan.8 High-risk (HR) HPV subtypes 16 and 18 have potential to transform epithelial cells which are derived from anogenital region. The transforming potential of HR-HPV is mainly due to the viral oncoproteins, E6 and E7, that deactivate tumor-suppressor proteins, p53 and pRb respectively. Expression of HR-HPV E6 and E7 causes cellular proliferation, dysregulated cell cycle, increased mutation and chromosomal instability.5
Minimally or non-invasive liquid biopsies including blood, cerebrospinal fluid, saliva and urine have recently gained much attention for diagnostic and prognostic purpose
in HNSCC.9 Despite the fact that the presence of HPV-16 in HNSCC is site-specific, studies have utilized either tumor tissue, saliva or plasma for HPV detection.5 While local
data on HR-HPV-16 in the subsets of HNSCC including oral/oropharyngeal squamous cell carcinoma are available, data on HNSCC including all subsites are lacking. Moreover,
data comparing blood and saliva for the presence of HPV-16 site-specificity in HNSCC are scarce. Therefore, we aimed to evaluate the frequency of HPV-16 in both blood and saliva in HNSCC in our population and also investigated its association with clinicopathological features.
METHODOLOGY
This observational study was conducted between September 2018 to August 2019 in Cancer Genetics and Molecular Cancer Therapeutics Lab at Quaid-i-Azam University Islamabad after approval from Institutional review board (IRB-QAU-170). A total of 200 samples comprising 100 blood and 100 saliva samples were collected. Of 200 samples, 100 samples (saliva, n = 50; blood, n = 50) were collected from 50 histopathologically confirmed head and neck squamous cell carcinoma patients who were admitted for surgical treatment at Pakistan Institute of Medical Sciences (PIMS), Islamabad. The rest of the 100 samples (saliva, n = 50, blood, n = 50) were collected from 50 healthy individuals without a history of cancer as controls who came for third molar extraction in same hospital. Both saliva and blood samples were collected from each patient and healthy individual. Patients receiving radio-chemotherapy, history of any other cancer and recurrent cases were excluded from the study. Medical records were retrieved for age, gender, marital status, medical and family history, habits and histopathological grade and written informed consent were obtained from patients and healthy individuals.
Sample collection
A 5 ml of blood and 5 ml of saliva were collected in vacutainer tubes containing ethylene-diamine-tetra-acetic acid (EDTA) (BD Vacutainer/EDTA-Franklin-Lakes-NJUSA) and falcon tubes respectively. The blood and saliva samples were stored at 4ºC and -20ºC respectively until DNA extraction.
DNA extraction
DNA extraction was performed using conventional phenol-chloroform method.10 Briefly, Eppendorf tube (1.5 ml) (Axygen-USA) containing 750-µl of blood (600 µl saliva) and 750 µl of RBCs lysis buffer (in blood samples only) was processed for centrifugation at 13,000 rpm for 1 minute followed by removal of supernatant, resuspension of pellet in 500 µl of RBCs lysis buffer and centrifugation at 13000 rpm for 60 seconds (saliva was centrifuged without adding RBCs lysis buffer). Consecutively, the supernatant was discarded followed by resuspension of pellet in WBCs lysis buffer (500 µl), sodium dodecyl sulfate solution (15 µl) and proteinase-K (10 µl) and incubation at 37 °C overnight (56 °C for 10 hours for saliva). Following incubation, 500 µl solution of freshly prepared phenol/chloroform/isoamyl alcohol in a ratio of 25 µl: 24 µl: 1 µl was added and processed for centrifugation at 13000 rpm for ten minutes. Following centrifugation, top layer was pipetted off into new Eppendorf tube and this step was repeated twice. Subsequently, sodium acetate (55 µl) and
cold iso-propanol (500 µl) were added to the aqueous layer in Eppendorf tube and DNA was precipitated by inverting the tube multiple times (in case of saliva, the tube was then
stored at -40 °C overnight) followed by centrifugation at 13,000 rpm for 10 minutes. The pellet was washed with 70% cold ethanol (200 µl), centrifuged and air-dried at 45 °C for 5 minutes in vacuum (Eppendorf-Hamburg-Germany). Finally, Tris-EDTA-buffer (150 µl) was used to dissolve DNA and incubated at 37°C overnight until further processing.
Quantification of DNA was determined by GenerayUV-photometer (Biometra-Goettingen-Germany) with 260 nanometer (nm) optical-density. The DNA integrity was assessed using 1% agarose-TBE stained with 10 µl ethidiumbromide via electrophoresis. DNA (5 µl) with 5 µl bromophenol blue dye was loaded in gel-wells and electrophoresed at 90 volts for 1.5 hours in 1X-buffer under UV-photometer (Biometra-Goettingen-Germany).
Primers design
Following forward (F) and reverse (R) primers were designed for HPV-16 genes E6 and E7 and human beta globin gene (BG) as internal control using Primer 3, Integrated DNA technologies. Human beta globin is the most commonly used gene to assess quantity of extracted DNA. The sequences of E6, E7 and human beta globin gene (internal control) were retrieved from Ensembl and then hits and alignments were checked with BLASTN and BLAT.
E6: forward 5’GCTGGGTTTCTCTACGTG3’ (62.1°C), reverse 5’AGCGACCCAGAAAGTTAC3’ (62.4 °C) and product-size of 430 base-pairs (bp) E7: forward 5’GGAGATACACCTACATTGC3’ (60.2°C), reverse 5’TTGTACGCACAACCGAAG3’ (63.3°C) and product-size of 203 bp BG: forward 5’AGATAGAGCCACTGACCCCA3’ (62.4°C), reverse 5’ACATCCCTCAATCTGCCAGG 3’ (63.3°C) and product-size of 268 bp.
Polymerase chain reaction (PCR) and gel electrophoresis
The 0.2 ml PCR-tube (Axygen-USA) was kept in heating block of the thermal cycler (Biometra-Tpersonal-GottingenGermany). Reaction mixture, with a 25 µl total reaction volume comprised of buffer 2.5µl (100M TrisHcl, 500mM KCL, 8.3 pH), 1.5 of MgCl2 (25mM), 1.5µl dNTPs (10mM, Fermantas-UK), 1µl of forward-reverse primers (0.1M), 0.6 µl of Taq-polymerase (Fermentas-UK), 14.9-15.4 µl of PCR water and 2-2.5 µl of DNA.
Thermocycler conditions for E6, E7 and BG were as follows; initial denaturation (96 °C, 5 minutes) followed by 40 denaturation cycles at 96 °C for 1 minute, annealing for 1 minute (E6 = 57 °C, E7 = 59 °C and BG = 61 °C) and initial and final extension at 72 °C for 1 and 10 minute/s respectively followed by analysis of amplified products on 2 % agarose TBE gel. The TBE gel was stained with 10 µl ethidium bromide followed by electrophoresis at 120-volts for 1.5 hour. Size of amplified product was determined using DNA-gene ladder of 100 base-pairs (MBI-Fermentas-LifeSciences-York-UK). From each PCR-reaction, 3 µl amplifiedsample mixed with 3 µl loading-buffer was loaded in wells. DNA fragments were visualized under UV-photometer (Biometra-Gottingen-Germany) and photographs were captured using digital camera (DC-290-Kodak-USA).
DATA ANALYSIS
The data were entered analyzed using SPSS version 24. The frequency and distribution of clinicopathological variables including gender, age, site, grade, ethnic group and smoking
habits were calculated in percentages. Chi-square test was used to determine the association of HPV-16 and clinicopathological variables. A P-value of less than 0.05 was considered as statistically significant.
RESULTS
The mean age of the HNSCC patients was 57.44 ± 14.98 years with age ranged from 25 to 85 years. Majority of the patients were in fifth and sixth decade. The mean age of controls was 54.92 ± 17.61 years with age ranged from 17 to 82 years. The clinicopathologic parameters and HPV-16 status of the HNSCC and controls are shown in Table 1. Next, the association of HPV-16 positivity in saliva and blood with clinicopathological characteristics was evaluated. No statistically significant association was found between HPV-16 positivity in saliva and blood and clinicopathological characteristics (P > 0.05) as shown in Table 2. A total of 200 extracted DNA (saliva; n = 100, blood; n = 100) were tested in polymerase chain reaction with beta-globin to investigate the presence of DNA prior to HPV-16 E6 and E7. Human beta-globin was successfully detected in all samples (HNSCC and healthy individuals) as shown in Figure 1. Of 100 samples (saliva; n = 50, blood; n = 50) from HNSCC patients, a total of 11 (11%) samples were positive
Table 1: Clinicopathologic parameters and HPV-16 status
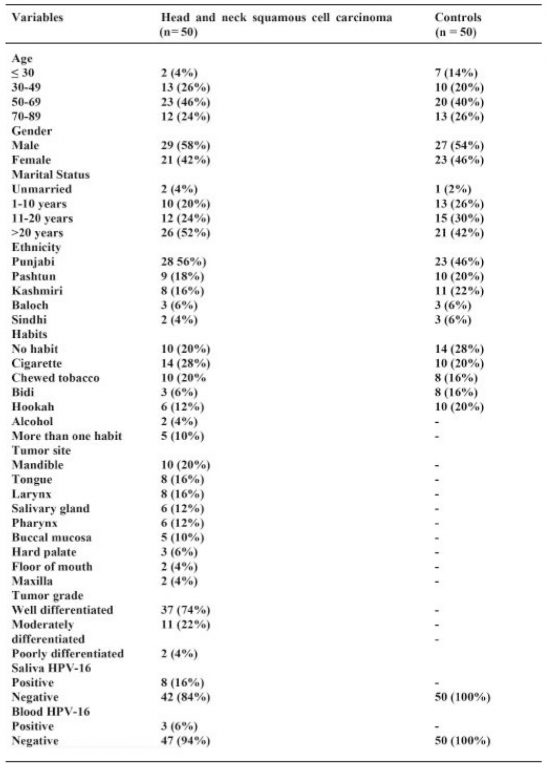
Table 2: Association of HPV-16 and head and neck squamous cell carcinoma
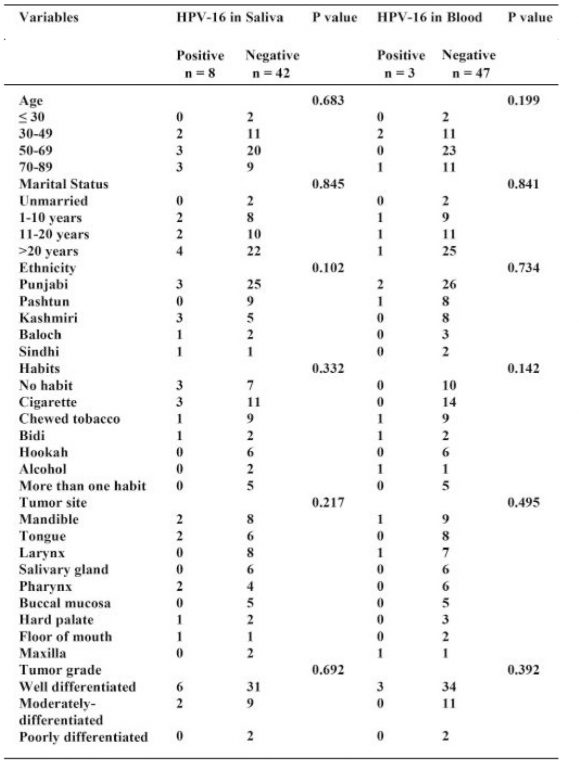
Figure 1: Electrophoretogram of 2% agarose gel showing amplified beta globin gene products of 268 bp in normal individuals and HNSCC patients
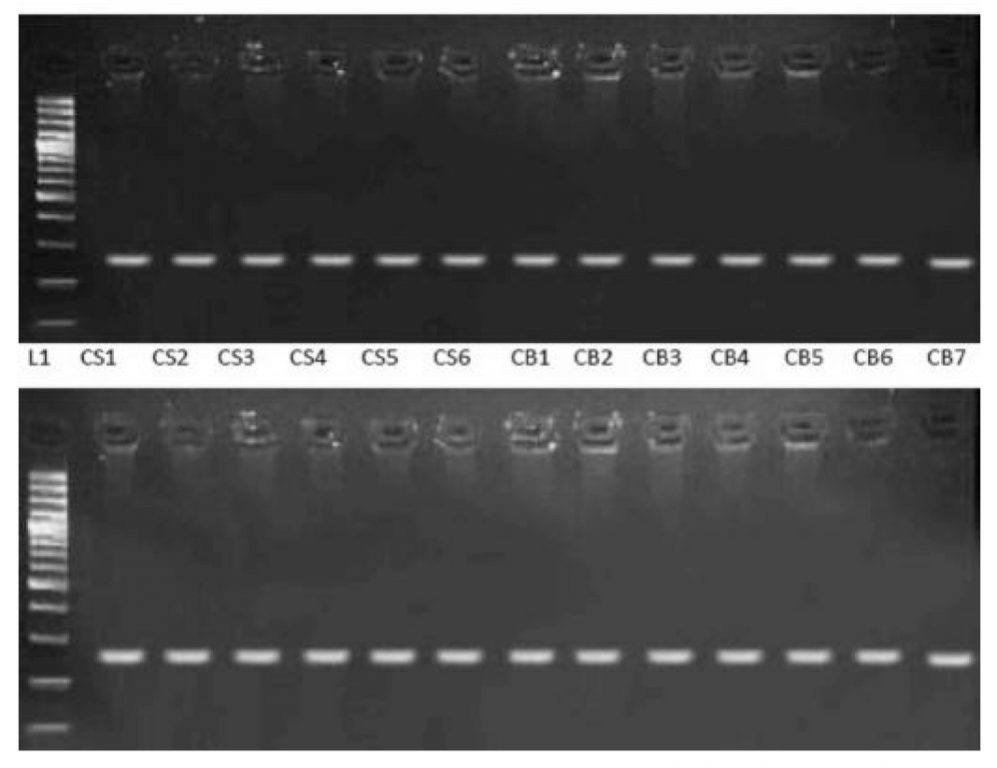
L1: Lane of 100 bp ladder, Lanes NS1-NS6: Saliva samples Lanes NB1-NB7: Blood samples, Lanes CS1-CS6: Saliva samples Lanes CB1-CB7: Blood samples
Figure 2: Electrophoretogram of 2% agarose gel showing amplified E6 products in HNSCC patients
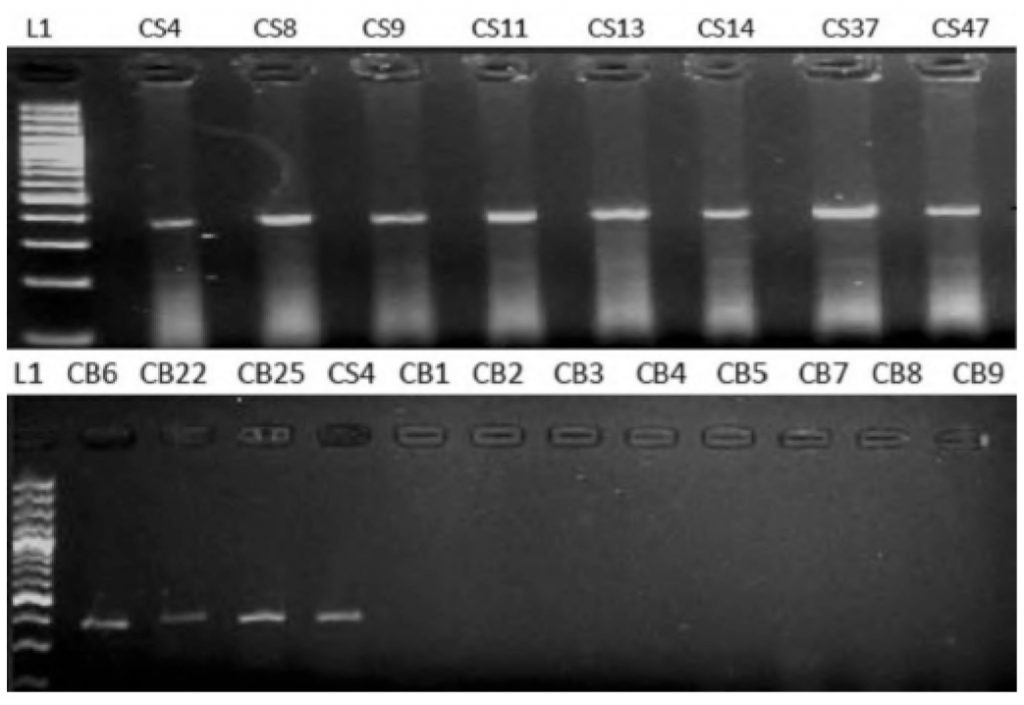
Lanes CS4, CS8, CS9, CS11, CS13, CS14, CS37 and CS47: Saliva samples, Lanes CB6, B22, B25: Blood samples
Figure 3: Electrophoretogram of 2% agarose gel showing no amplified E7 gene products in HNSCC
Lanes CS1-CS12: Saliva samples, Lanes CB1-CB12: Blood samples
Figure 4: Electrophoretogram of 2% agarose gel showing no amplified E6/E7 gene in normal individuals
Lanes NS1-NS7: Saliva samples, Lanes NB1-NB6: Blood samples
for HPV-16 E6 gene. Of 11 HPV-16 positive samples, 8 (16%) were detected in saliva (n = 50) and 3 (6%) were detected in plasma (n = 50) as shown in Figure 2 and none of the HNSCC samples were positive for HPV-16 E7 gene as shown in Figure 3. Of 100 samples (saliva, n = 50; blood, n = 50) from control group, none of the samples were positive for E6/E7 (Figure 4).
DISCUSSION
In our study, no HPV-16 was observed in both saliva and blood samples in control patients. Our findings are in contrast to Khayani et al. who reported salivary-HPV-16 in 03/35 controls and Gichki et al. who found HPV-16 in 4/200 normal oral tissue scrapings.11,12 The difference in our findings could be due to techniques employed in detecting HPV. We
utilized conventional PCR which is less sensitive in detecting viral-DNA compared to quantitative real-time or nested PCR. However, in accordance with our results, literature also reported negligible or low prevalence of HR-HPV in saliva in healthy individuals indicating the absence of HRHPV-16 in non-HNSCC patients.13,14
Our study showed 11% HPV-16 positivity in HNSCC patients. Furthermore, HPV-16 was detected more in saliva (16%) than blood (6%) particularly in squamous cell carcinomas arising from oropharynx (mandible, tongue, palate, floor of the mouth and pharynx) suggesting that detection of HPV-16 in saliva is site-specific. Our findings are in agreement with previous studies.15,16,17,18 The existence of HPV-16 in saliva is due to the infected cells generating viral particles or the viruses releasing from the dead cells into saliva.15 In contrast, Wang et al. reported 40% (n = 30) and 86% (n = 21) prevalence rates of HPV-16 in saliva and plasma respectively in HNSCC patients. Moreover, they also reported that detection of HPV-16 in plasma was not site-specific and concluded plasma as an optimal biofluid for detecting high-risk HPV-16 in HNSCC.19
In the present study, the smoking rate (46%) in a form of cigarette, bidi and hookah was high compared to chewed tobacco and alcohol in HNSCC patients, however, no significant association was found between overall use of tobacco and HPV-16 which is in agreement with previous study.20 Our data indicate that both tobacco and HPV-16 are independent risk factors for tumorigenesis of HNSCC in Punjab (the largest province of Pakistan). In contrast, previous study reported significant association between HPV-16 and chewed tobacco users.21 The plausible reasons for the difference in findings could be related to sample size (100 oral squamous cell carcinomas compared to 50 HNSCC cases in the current study). Furthermore, we couldn’t find statistically significant association between HPV-16 and age and gender which is in accordance with previous studies, however, others reported significant association of male predominance in HPV-associated HNSCC.20,22,23 Additionally, we couldn’t establish statistically significant association between HPV-16 and ethnicity suggesting that HPV-16 is independent of ethnic diversity in Pakistani population. Microscopically, most of the HNSCC cases present with moderate-poorly differentiated squamous cell carcinoma. The moderately-differentiated are observed with HPVnegative while poorly differentiated are associated with HPV-positive HNSCC, however, no such association was observed in our study.20,24,25 While our findings are consistent with many of the reported studies in literature, it is arduous to compare various
local published studies on HPV-16 mainly due to the wide discrepancy in findings. Plausible reasons of this discrepancy are variations in sample size, type of sample used for instance tissue, saliva, oral scraping or saliva-rinse, techniques employed for HPV-16 detection and studies involving only one subset of HNSCC mainly oral or oropharyngeal squamous-cell-carcinoma.
LIMITATIONS
Detection of HPV-16 in both saliva and plasma (liquid biopsy) with diversified ethnic background and HNSCC subsites was the strength of our study. However, our study had few limitations including small sample size, use of conventional PCR instead of more sensitive techniques due to limited available resources, detection of one HPV-subtype and lack of data on sexual habits due to social and ethical issues.
CONCLUSION
Within the limit of current study, HPV-16 has a low-frequency in the saliva and blood of HNSCC in our population. Moreover, HPV-16 was detected more in saliva than blood particularly in squamous cell carcinomas arising from oropharynx suggesting that detection of HPV-16 in saliva is site-specific. Further studies on HNSCC with large sample including all subsites of squamous cell carcinoma arising in head and neck region, liquid biopsy, other HPV-subtypes, tobacco and non-tobacco users are recommended for future research.
ACKNOWLEDGMENT
The authors are grateful to the staff at Cancer Genetics and Molecular Cancer Therapeutics Lab, Quaid-i-Azam University Islamabad for their help and cooperation and Asif Hanif for statistical analysis.
CONFLICT OF INTEREST
The authors declare no conflict of interest ETHICAL APPROVAL All procedures performed were in accordance with the ethical standards of the institutional research committee and conformed with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
REFERENCES
- Jou A, Hess J. Epidemiology and molecular biology of head and neck cancer. Oncol Res Treat. 2017;40:328-332. https://doi.org/10.1159/000477127
- Perdomo S, Roa GM, Brennan P, Forman D, Sierra MS. Head and neck cancer burden and preventive measures in Central and South America. Cancer Epidemiol. 2016;44:S43-52. https://doi.org/10.1016/j.canep.2016.03.012
- Qureshi MA, Mirza T, Khan S, Sikandar B, Zahid M, Aftab M, et al. Cancer patterns in Karachi (all districts), Pakistan: first results (2010-2015) from a pathology-based cancer registry of the largest government-run diagnostic and reference center of Karachi. Cancer Epidemiol. 2016;44:114-122. https://doi.org/10.1016/j.canep.2016.08.011
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. https://doi.org/10.3322/caac.21492
- Farsi NJ, Rousseau MC, Schlecht N, Castonguay G, Allison P, Nguyen-Tan PF, et al. Aetiological heterogeneity of head and neck squamous cell carcinomas: the role of human papillomavirus infections, smoking and alcohol. Carcinogenesis. 2017;38:1188-1195. https://doi.org/10.1093/carcin/bgx106
- Devaraja K, Aggarwal S, Verma SS, Gupta SC. Clinico-pathological peculiarities of human papilloma virus driven head and neck squamous cell carcinoma: A comprehensive update. Life Sci. 2020;245:117383. https://doi.org/10.1016/j.lfs.2020.117383
- Gotz C, Bischof C, Wolff KD, Kolk A. Detection of HPV infection in head and neck cancers: Promise and pitfalls in the last ten years: A meta-analysis. Mol Clin Oncol. 2019; 10:17-28. 10.3892/mco.2018.1749
- Bukhari N, Joseph JP, Hussain SS, Khan MA, Wakim MJ, Yahya EB, et al. Prevalence of human papilloma virus sub genotypes following head and neck squamous cell carcinomas in asian continent, a systematic review article. Asian Pac J Cancer Prev. 2019;20:3269-3277. https://doi.org/10.31557/APJCP.2019.20.11.3269
- Spector ME, Farlow JL, Haring CT, Brenner JC, Birkeland AC. The potential for liquid biopsies in head and neck cancer. Discov Med. 2018;25:251-257.
- Sambrook J, Russell DW. Purification of nucleic acids by extraction with phenol:chloroform. CSH Protoc. 2006;2006:pdb.prot4455. https://doi.org/10.1101/pdb.prot4455
- Khyani IA, Qureshi MA, Mirza T, Farooq MU. Salivary detection of human Papilloma virus 16 & 18 in pre-malignant and malignant lesions of oral cavity: is it feasible in Pakistani context of socio-cultural taboos? Pak J Med Sci. 2015;31:1104-1109. https://doi.org/10.12669/pjms.315.7093
- Gichki AS, Buajeeb W, Doungudomdacha S, Khovidhunkit SO. Detection of human papillomavirus in normal oral cavity in a group of Pakistani subjects using real-time PCR. Asian Pac J Cancer Prev. 2012;13:2299-304. https://doi.org/10.7314/APJCP.2012.13.5.2299
- Kreimer AR, Bhatia RK, Messeguer AL, Gonz¡lez P, Herrero R, Giuliano AR. Oral human papillomavirus in healthy individuals: systematic review of the literature. Sex Transm Dis. 2010;37:386-391. https://doi.org/10.1097/OLQ.0b013e3181c94a3b
- Gillison ML, Broutian T, Pickard RK, Tong ZY, Xiao W, Kahle L et al. Prevalence of oral HPV infection in the United States, 2009- 2010. JAMA. 2012;307:693-703. https://doi.org/10.1001/jama.2012.101
- Wasserman JK, Rourke R, Purgina B, Caulley L, Dimitroulakos J, Corsten M, et al. Correction to: HPV DNA in saliva from patients with SCC of the head and neck is specific for p16-positive oropharyngeal tumours. J Otolaryngol Head Neck Surg. 2018;47:49. https://doi.org/10.1186/s40463-018-0294-7
- Fauzi FH, Hamzan NI, Rahman NA, Mohamad I, Suraiya S, Kallarakkal TG, Mohamad S. Detection of human papillomavirus types 16 and 18 in oral squamous cell carcinoma samples in Malaysia. Arch Orofac Sci. 2019;14:21-29.
- Parshad S, Nandi S, Marwah N, Mehta P, Tripathi M, Netrapal et al. Human papillomavirus 16 and 18 in squamous cell carcinoma of oral cavity and sexual practices: A pilot study at a Tertiary Care Hospital of North India. Natl J Maxillofac Surg. 2015;6:185-189. https://doi.org/10.4103/0975-5950.183857
- Awan MS, Irfan B, Zahid I, Mirza Y, Ali SA. Comparison of polymerase chain reaction and immunohistochemistry assays for analysing human papillomavirus infection in oral squamous cell carcinoma. J Clin Diagn Res. 2017;11:XC10-13. https://doi.org/10.7860/JCDR/2017/24742.10119
- Wang Y, Springer S, Mulvey CL, Silliman N, Schaefer J, Sausen M, et al. Detection of somatic mutations and HPV in the saliva and plasma of patients with head and neck squamous cell carcinomas. Sci Transl Med. 2015;7:293ra104.
- Khasawneh AI, Himsawi N, Abu-Raideh J, Salameh M, Abdullah N, Khasawneh R. Prevalence of human papillomavirus associated with head and neck squamous cell carcinoma in Jordanian patients. Open Microbiol J [Internet]. 2020 April [cited 2020 May 7];14:57-64. Available from: https://benthamopen.com/FULLTEXT/TOMICROJ14-57. https://doi.org/10.2174/1874285802014010057
- Zil-E-Rubab, Baig S, Zaman U, Lucky MH. Human papilloma virus 16/18: Fabricator of trouble in oral squamous cell carcinoma. Int J Infect Dis. 2018;69:115-119. https://doi.org/10.1016/j.ijid.2018.02.003
- Adnan Ali SM, Awan MS, Atif S, Ali N, Mirza Y. Correlation of human papillomavirus infection and clinical parameters with five-year survival in oral squamous cell carcinoma. J Laryngol Otol. 2018;132:628-635. https://doi.org/10.1017/S0022215118000361
- Fakhry C, Krapcho M, Eisele DW, D’Souza G. Head and neck squamous cell cancers in the United States are rare and the risk now is higher among white individuals compared with black individuals. Cancer. 2018;124:2125-2133. https://doi.org/10.1002/cncr.31322
- Hennessey PT, Westra WH, Califano JA. Human papillomavirus and head and neck squamous cell carcinoma: Recent evidence and clinical implications. J Dent Res. 2009; 88:300-306. https://doi.org/10.1177/0022034509333371
- Abu Qatouseh L, Sabri I, Alkhatib I, Atwa E, Arafat T. Detection of high-risk human papillomavirus genotypes 16 and 18 in head and neck squamous cell carcinomas in Jordan. Asian Pac J Cancer Prev. 2017; 18:1337-1341.

